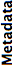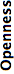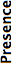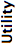Supported Formats¶
Ratings legend and definitions
You can sort this table by clicking on any of the headings.
Bio-Formats currently supports 147 formats
- Ratings legend and definitions

Outstanding 
Very good 
Good 
Fair 
Poor - Pixels
- Our estimation of Bio-Formats’ ability to reliably extract complete and accurate pixel values from files in that format. The better this score, the more confident we are that Bio-Formats will successfully read your file without displaying an error message or displaying an erroneous image.
- Metadata
- Our certainty in the thoroughness and correctness of Bio-Formats’ metadata extraction and conversion from files of that format into standard OME-XML. The better this score, the more confident we are that all meaningful metadata will be parsed and populated as OME-XML.
- Openness
- This is not a direct expression of Bio-Formats’ performance, but rather indicates the level of cooperation the format’s controlling interest has demonstrated toward the scientific community with respect to the format. The better this score, the more tools (specification documents, source code, sample files, etc.) have been made available.
- Presence
- This is also not directly related to Bio-Formats, but instead represents our understanding of the format’s popularity, and is also as a measure of compatibility between applications. The better this score, the more common the format and the more software packages include support for it.
- Utility
- Our opinion of the format’s suitability for storing metadata-rich microscopy image data. The better this score, the wider the variety of information that can be effectively stored in the format.
- Export
- This indicates whether Bio-Formats is capable of writing the format (Bio-Formats can read every format on this list).
- BSD
- This indicates whether format is BSD-licensed. By default, format readers and writers are GPL-licensed.
- Multiple Images
- This indicates whether the format can store multiple Images (in OME-XML terminology) or series (in Bio-Formats API terminology).
- Pyramid
- This indicates whether the format can store a single image at multiple resolutions, typically referred to as an image pyramid.