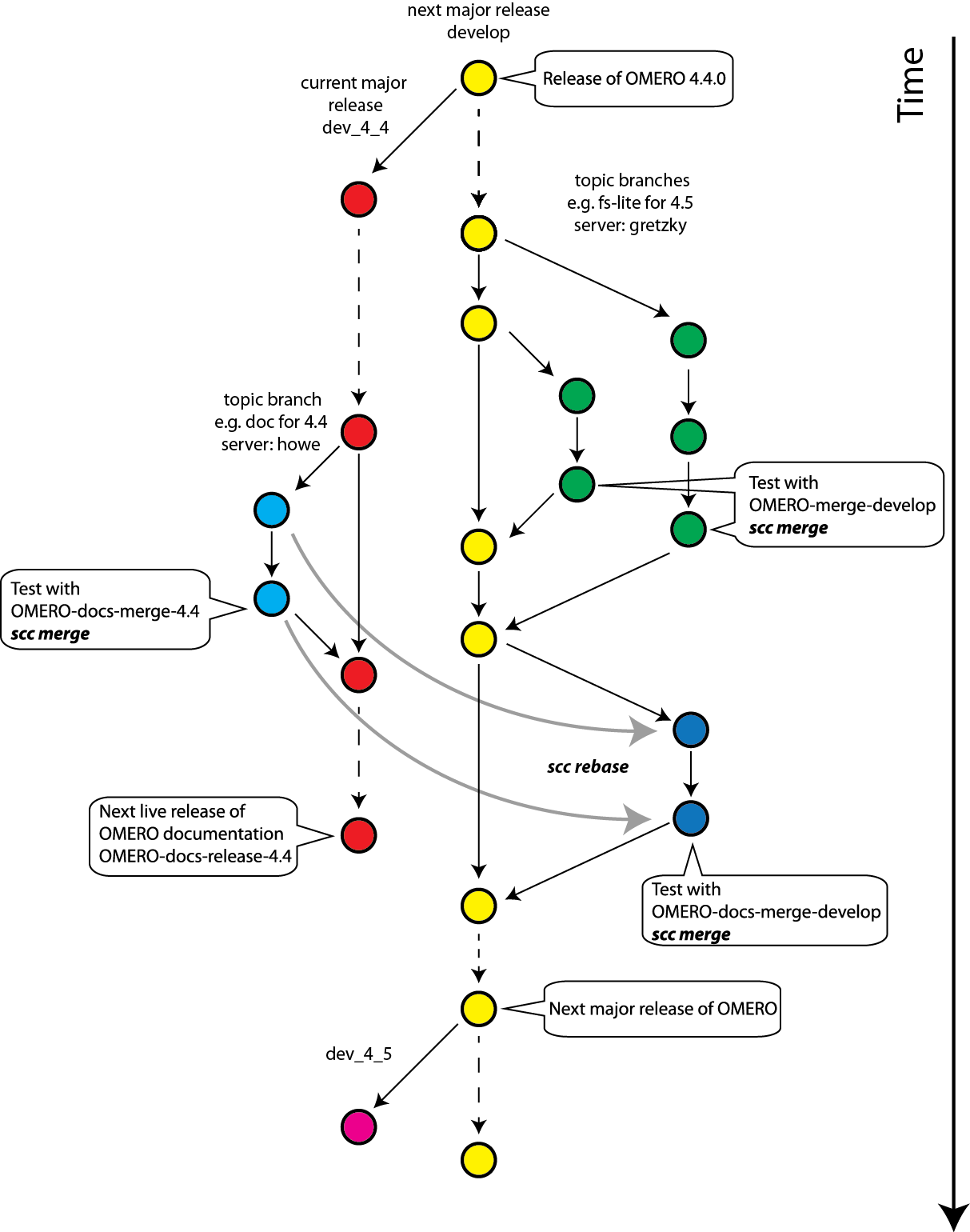
Continuous integration workflow¶
Definitions¶
Versioning¶
Development branches¶
Most of the OME code is split between four repositories: openmicroscopy.git, bioformats.git, scripts.git, ome-documentation.git. For each repository, two development branches are maintained simultaneously:
- The “dev_4_4” branch (“stable” branch) contains work on the stable release version and will only be released as a point release.
- The “develop” branch contains work on the next major release.
It is possible to have a point release immediately, while still working on more major releases by ensuring that (nearly) all commits that are applied to dev_4_4 are applied to develop in order to prevent regressions.
Labels¶
Labels are applied to PRs on GitHub under the “Issues” tab of each repository.
The next point release consists of PRs labelled with “dev_4_4”, which is the name of the branch which they will be merged into.
The next major release consists of PRs labelled with “develop”, which is also the name of the branch which they will be merged into.
Three labels are used in the PR reviewing process: “include”, “exclude”, and “on hold”. The “include” label allows you to include a PR opened by a non-member of the OME organization in the merge builds for review. The “exclude” label allows you to exclude a PR opened by any user from the merge builds. The “on hold” label allows you to signal that a PR should not be reviewed or merged, even though it is not excluded.
OMERO jobs¶
| Job task | Next point release | Next major release |
|---|---|---|
| Build OMERO using Ice 3.3 | OMERO-stable | OMERO-trunk |
| Build OMERO using Ice 3.4 | OMERO-stable-ice34 | OMERO-trunk-ice34 |
| Build an OMERO Virtual Appliance | OMERO-stable-virtualbox | OMERO-trunk-virtualbox |
| Review OMERO PRs using Ice 3.3 | OMERO-merge-stable | OMERO-merge-develop |
| Review OMERO PRs using Ice 3.4 | OMERO-merge-stable-ice34 | OMERO-merge-develop-ice34 |
| Review OMERO PRs using a Virtual Appliance | OMERO-merge-stable-virtualbox | OMERO-merge-develop-virtualbox |
| Update submodules | OMERO-submods-stable | OMERO-submods-develop |
Next point release¶
The branch for the next point release of OMERO is dev_4_4. All jobs are listed under the Stable view tab of Jenkins.
- OMERO-stable
This job is used to build the dev_4_4 branch of OMERO with Ice 3.3
- Builds the OMERO.server and the clients using OMERO.sh
- Archives the build artifacts
- If the build is promoted, copies the artifacts to necromancer
- OMERO-stable-ice34
This job is used to build the dev_4_4 branch of OMERO with Ice 3.4
- Builds the OMERO.server and the clients using OMERO.sh
- Archives the build artifacts
- If the build is promoted, copies the artifacts to necromancer
- OMERO-stable-virtualbox
This job is used to build a Virtual Appliance from the dev_4_4 branch of OMERO
- Builds a VM using omerovm.sh
- OMERO-merge-stable
This job is used to review the PRs opened against the dev_4_4 branch of OMERO with Ice 3.3
- OMERO-merge-stable-ice34
This job is used to review the PRs opened against the dev_4_4 branch of OMERO with Ice 3.4
- Checks out the merge/dev_4_4/latest branch of the snoopycrimecop fork of openmicroscopy.git
- Builds the OMERO.server and the clients using OMERO.sh
- Archives the build artifacts
- OMERO-merge-stable-virtualbox
This job is used to build a Virtual Appliance from the dev_4_4 branch with the PRs opened against the dev_4_4 branch of OMERO
- Checks out the merge/dev_4_4/latest branch of the snoopycrimecop fork of openmicroscopy.git
- Builds a VM using omerovm.sh
- OMERO-submods-stable
This job is used to update the submodules on the dev_4_4 branch
- Updates submodules using scc update-submodules and pushes the merged branch to snoopycrimecop/merge/dev_4_4/submodules
- If the submodules are updated, opens a new PR or updates the existing dev_4_4 submodules PR
Next major release¶
The branch for the next major release of OMERO is develop. All jobs are listed under the Trunk view tab of Jenkins.
- OMERO-trunk
This job is used to build the develop branch of OMERO with Ice 3.3.
- Builds the OMERO.server and the clients using OMERO.sh
- Archives the build artifacts
- If the build is promoted, copies the artifacts to necromancer
- OMERO-trunk-ice34
This job is used to build the develop branch of OMERO with Ice 3.4
- Builds the OMERO.server and the clients using OMERO.sh
- Archives the build artifacts
- If the build is promoted, copies the artifacts to necromancer
- OMERO-trunk-virtualbox
This job is used to build a Virtual Appliance from the develop branch of OMERO
- Builds a VM using omerovm.sh
- OMERO-merge-develop
This job is used to review the PRs opened against the develop branch of OMERO with Ice 3.3
- OMERO-merge-develop-ice34
This job is used to review the PRs opened against the develop branch of OMERO with Ice 3.4
- Checks out the merge/develop/latest branch of the snoopycrimecop fork of openmicroscopy.git
- Builds the OMERO.server and the clients using OMERO.sh
- Archives the build artifacts
- OMERO-merge-develop-virtualbox
This job is used to build a Virtual Appliance from the develop branch with the PRs opened against the develop branch of OMERO
- Checks out the merge/develop/latest branch of the snoopycrimecop fork of openmicroscopy.git
- Builds a VM using omerovm.sh
- OMERO-submods-develop
This job is used to update the submodules on the develop branch
- Updates submodules using scc update-submodules and pushes the merge branch to snoopycrimecop/merge/develop/submodules
- If the submodules are updated, opens a new PR or updates the existing dev_4_4 submodules PR
Bio-Formats jobs¶
All jobs are listed under the Bio-Formats view tab of Jenkins.
| Job task | Next point release | Next major release |
|---|---|---|
| Build Bio-Formats | BIOFORMATS-stable | BIOFORMATS-trunk |
| Publish Bio-Formats to the LOCI Nexus repository | BIOFORMATS-maven-stable | BIOFORMATS-maven |
| Review Bio-Formats PRs | BIOFORMATS-merge-stable | BIOFORMATS-merge-develop |
| Run automated tests against directories on squig | BIOFORMATS-full-repository-stable | BIOFORMATS-full-repository-develop |
Next point release¶
The branch for the next point release of Bio-Formats is dev_4_4.
- BIOFORMATS-stable
This job is used to build the dev_4_4 branch of Bio-Formats
- Builds Bio-Formats using ant clean jars tools tools-ome utils dist-bftools
- Runs Bio-Formats tests using ant test-common test-ome-xml test-formats test-ome-io
- BIOFORMATS-maven-stable
- This job is used to publish the dev_4_4 branch of Bio-Formats to the LOCI Nexus repository
- BIOFORMATS-merge-stable
This job is used to review the PRs opened against the dev_4_4 branch of Bio-Formats by running basic unit tests, checking for open file handles, and checking for regressions across a representative subset of the data repository
- Merges PRs using scc merge
- Builds Bio-Formats using ant clean jars tools tools-ome utils dist-bftools
- Runs Bio-Formats full test-suite using ant test
- BIOFORMATS-full-repository-stable
This job is used to review the PRs opened against the dev_4_4 branch of Bio-Formats by running automated tests against directories on squig
- Merges PRs using scc merge
- Run tests against directories configured by --test dirname comments on the PRs
Next major release¶
The branch for the next major release of Bio-Formats is develop.
- BIOFORMATS-trunk
This job is used to build the develop branch of Bio-Formats
- Builds Bio-Formats using ant clean jars tools tools-ome utils dist-bftools
- Runs Bio-Formats tests using ant test-common test-ome-xml test-formats test-ome-io
- BIOFORMATS-maven
- This job is used to publish the develop branch of Bio-Formats to the LOCI Nexus repository
- BIOFORMATS-merge-develop
This job is used to review the PRs opened against the develop branch of Bio-Formats by running basic unit tests, checking for open file handles, and checking for regressions across a representative subset of the data repository
- Merges PRs using scc merge
- Builds Bio-Formats using ant clean jars tools tools-ome utils dist-bftools
- Runs Bio-Formats full test-suite using ant test
- BIOFORMATS-full-repository-develop
This job is used to review the PRs opened against the develop branch of Bio-Formats by running automated tests against directories on squig
- Merges PRs using scc merge
- Run tests against directories configured by --test dirname comments on the PRs
Documentation jobs¶
All documentation jobs are listed under the Docs view tab of Jenkins.
| Job task | Current release/next point release | Next major release |
|---|---|---|
| Publish OMERO documentation | OMERO-docs-release-stable | OMERO-docs-release-develop |
| Publish Bio-Formats documentation | BIOFORMATS-docs-release-stable | BIOFORMATS-docs-release-develop |
| Publish OME Model documentation | FORMATS-docs-release-stable | FORMATS-docs-release-develop |
| Review OMERO documentation PRs | OMERO-docs-merge-stable | OMERO-docs-merge-develop |
| Review Bio-Formats documentation PRs | BIOFORMATS-docs-merge-stable | BIOFORMATS-docs-merge-develop |
| Review OME Model documentation PRs | FORMATS-docs-merge-stable | FORMATS-docs-merge-develop |
Configuration¶
For all jobs building documentation using Sphinx, the HTML documentation theme hosted at https://github.com/openmicroscopy/sphinx_theme repository is copied to the relevant themes/ folder. The following environment variables are then used:
- the Sphinx building options, SPHINXOPTS, is set to -W -D html_theme=sphinx_theme,
- the release number of the documentation is set by OMERO_RELEASE, BF_RELEASE or FORMATS_RELEASE,
- the source code links use SOURCE_USER and SOURCE_BRANCH,
- for the Bio-Formats and OMERO sets of documentation, the name of the Jenkins job is set by JENKINS_JOB.
Current release/next point release¶
The branch for the current release of OMERO/Bio-Formats/OME model is dev_4_4.
- OMERO-docs-release-stable
This job is used to build the dev_4_4 of the OMERO documentation and publish the official documentation for the current release of OMERO
Runs make clean html latexpdf to build the HTML and PDF versions of the Sphinx documentation
Runs make linkcheck and parse the Sphinx linkcheck output
- If the build is promoted,
- Copies the HTML and PDF documentation over SSH to /var/www/www.openmicroscopy.org/sphinx-docs/omero-stable-release.tmp
- Runs scc deploy to update http://www.openmicroscopy.org/site/support/omero4/
- BIOFORMATS-docs-release-stable
This job is used to build the dev_4_4 of the Bio-Formats documentation and publish the official documentation for the current release of Bio-Formats
Runs make clean html latexpdf to build the HTML and PDF versions of the Sphinx documentation
Runs make linkcheck and parse the Sphinx linkcheck output
- If the build is promoted,
- Copies the HTML and PDF documentation over SSH to /var/www/www.openmicroscopy.org/sphinx-docs/bf-stable-release.tmp
- Runs scc deploy to update http://www.openmicroscopy.org/site/support/bio-formats4/
- FORMATS-docs-release-stable
This job is used to build the dev_4_4 of the OME Model documentation and publish the official documentation for the current release of Bio-Formats
Runs make clean html latexpdf to build the HTML and PDF versions of the Sphinx documentation
Runs make linkcheck and parse the Sphinx linkcheck output
- If the build is promoted,
- Copies the HTML and PDF documentation over SSH to /var/www/www.openmicroscopy.org/sphinx-docs/formats-stable-release.tmp
- OMERO-docs-merge-stable
This job is used to review the PRs opened against the dev_4_4 branch of the OMERO documentation
- Merges PRs using scc merge
- Runs make clean html latexpdf to build the HTML and PDF versions of the Sphinx documentation
- Runs make linkcheck and parse the Sphinx linkcheck output
- Copies the HTML and PDF documentation over SSH to /var/www/www.openmicroscopy.org/sphinx-docs/omero-stable-staging.tmp
- Runs scc deploy to update http://www.openmicroscopy.org/site/support/omero4-staging/
- BIOFORMATS-docs-merge-stable
This job is used to review the PRs opened against the dev_4_4 branch of the Bio-Formats documentation
- Merges PRs using scc merge
- Runs make clean html latexpdf to build the HTML and PDF versions of the Sphinx documentation
- Runs make linkcheck and parse the Sphinx linkcheck output
- Copies the HTML and PDF documentation over SSH to /var/www/www.openmicroscopy.org/sphinx-docs/bf-stable-staging.tmp
- Runs scc deploy to update http://www.openmicroscopy.org/site/support/bio-formats4-staging/
- FORMATS-docs-merge-stable
This job is used to review the PRs opened against the dev_4_4 branch of the OME Model documentation
- Merges PRs using scc merge
- Runs make clean html latexpdf to build the HTML and PDF versions of the Sphinx documentation
- Runs make linkcheck and parse the Sphinx linkcheck output
- Copies the HTML and PDF documentation over SSH to /var/www/www.openmicroscopy.org/sphinx-docs/formats-stable-staging.tmp
Next major release¶
The branch for the next major release of OMERO/Bio-Formats/OME model is develop.
- OMERO-docs-release-develop
This job is used to build the develop branch of the OMERO documentation and publish the official documentation for the next major release of OMERO
Runs make clean html latexpdf to build the HTML and PDF versions of the Sphinx documentation
Runs make linkcheck and parse the Sphinx linkcheck output
- If the build is promoted,
- Copies the HTML and PDF documentation over SSH to /var/www/www.openmicroscopy.org/sphinx-docs/omero-develop-release.tmp
- Runs scc deploy to update http://www.openmicroscopy.org/site/support/omero5/
- BIOFORMATS-docs-release-develop
This job is used to build the develop branch of the Bio-Formats documentation and publish the official documentation for the next major release of Bio-Formats
Runs make clean html latexpdf to build the HTML and PDF versions of the Sphinx documentation
Runs make linkcheck and parse the Sphinx linkcheck output
- If the build is promoted,
- Copies the HTML and PDF documentation over SSH to /var/www/www.openmicroscopy.org/sphinx-docs/bf-develop-release.tmp
- Runs scc deploy to update http://www.openmicroscopy.org/site/support/bio-formats5/
- FORMATS-docs-release-develop
This job is used to build the develop branch of the OME Model documentation and publish the official documentation
- Merges PRs using scc merge
- Runs make clean html latexpdf to build the HTML and PDF versions of the Sphinx documentation
- Runs make linkcheck and parse the Sphinx linkcheck output
- Copies the HTML and PDF documentation over SSH to /var/www/www.openmicroscopy.org/sphinx-docs/formats-develop-release.tmp
- Runs scc deploy to update http://www.openmicroscopy.org/site/support/ome-model/
- OMERO-docs-merge-develop
This job is used to review the PRs opened against the develop branch of the OMERO documentation
- Merges PRs using scc merge
- Runs make clean html latexpdf to build the HTML and PDF versions of the Sphinx documentation
- Runs make linkcheck and parse the Sphinx linkcheck output
- Copies the HTML and PDF documentation over SSH to /var/www/www.openmicroscopy.org/sphinx-docs/omero-develop-staging.tmp
- Runs scc deploy to update http://www.openmicroscopy.org/site/support/omero5-staging/
- BIOFORMATS-docs-merge-develop
This job is used to review the PRs opened against the develop branch of the Bio-Formats documentation
- Merges PRs using scc merge
- Runs make clean html latexpdf to build the HTML and PDF versions of the Sphinx documentation
- Runs make linkcheck and parse the Sphinx linkcheck output
- Copies the HTML and PDF documentation over SSH to /var/www/www.openmicroscopy.org/sphinx-docs/bf-develop-staging.tmp
- Runs scc deploy to update http://www.openmicroscopy.org/site/support/bio-formats5-staging/
- FORMATS-docs-merge-develop
This job is used to review the PRs opened against the develop branch of the OME Model documentation
- Merges PRs using scc merge
- Runs make clean html latexpdf to build the HTML and PDF versions of the Sphinx documentation
- Runs make linkcheck and parse the Sphinx linkcheck output
- Copies the HTML and PDF documentation over SSH to /var/www/www.openmicroscopy.org/sphinx-docs/formats-develop-staging.tmp
- Runs scc deploy to update http://www.openmicroscopy.org/site/support/ome-model-staging/
Linkcheck output parser¶
The output.txt file generated by Sphinx linkcheck builder is parsed using the Warnings Plugin. Depending on the nature of the links, warnings are generated as described in the following table:
| Type | Error code | Priority |
|---|---|---|
| local | High | |
| broken | HTTP Error 404 | Normal |
| broken | Anchor not found | Normal |
| broken | HTTP Error 403 | Low |
The build is marked as FAILED or UNSTABLE if the number of warnings of a given category exceeds a threshold. The table below lists the thresholds used for all the documentation builds:
| Priority | FAILED | UNSTABLE |
|---|---|---|
| High | 0 | |
| Normal | 0 | |
| Low | 10 |
Schematic drawing¶