The OME-XML format¶
OME-XML is a project to provide support for the exchange of microscope imaging data within the field of life sciences. Its key aspects include support for collaborative sharing of data between labs who may be using different apparatus, and the safeguarding of crucial metadata associated with image files, that can be lost when they are moved across systems (or in many cases, never recorded at all).
The term “OME-XML” applies to two interrelated but distinct things:
- A data model for representing microscopy metadata, for use with microscopy file formats, expressed as an XML schema.
- A file format for storing microscopy information (both pixels and metadata) using the OME-XML data model.
Note
OME-XML as a file format is somewhat superseded by OME-TIFF, which is the preferred container format for OME-XML encoded metadata.
The purpose of OME-XML is to provide a rich, extensible way to save information concerning microscopy experiments and the images acquired therein, including:
- dimensional parameters defining the scope of the image pixels (e.g. resolution, number of focal planes, number of time points, number of channels)
- the hardware configuration used to acquire the image planes (e.g. microscope, detectors, lenses, filters)
- the settings used with said hardware (e.g. physical size of the image planes in microns, laser gain and offset, channel configuration)
- the person performing the experiment
- some details on the experiment itself, such as a description, what type of experiment (e.g. FRET, time lapse, fluorescence lifetime) and events occurring during the acquisition (e.g. laser ablation, stage motion)
- additional custom information you may wish to provide about your experiment in a structured form (known as Structured Annotations)
Features and applications¶
The OME-XML file serves as a convenient file format for data migration from one site or user to another. The OME-XML file captures all image acquisition and experimental metadata, along with the binary image data, and packages it into an easily readable package. The paper describing the design and implementation of the OME-XML file appeared in Genome Biology.
Note
OME-XML files can be read by potentially any software package - you do not need OME image management software to use OME-XML.
Some specific features of the OME-XML file format:
- OME-XML files may contain one or more sets of 5-D pixels, for example raw data from a microscope, the deconvolved data, and a volume rendered view.
- OME-XML files contain all the metadata associated with an image, including the experimental (e.g. cells, genes) and acquisition (e.g. microscope light sources, filters, detectors) metadata.
- OME-XML Image pixels may be stored compressed directly in XML with base64 encoding. Compression of the pixels and the metadata is supported through widely-available patent-free compression schemes (gzip and bzip2). OME-XML Images are addressable by plane.
- OME-XML files have a built-in mechanism for supporting arbitrary user-defined data that can be used globally or attached to Images, Features (objects inside Images), and Datasets. Mechanisms for OME-compliant systems to populate databases with these user-defined fields is part of the specification. See the Structured Annotations XML Schema section.
The OME-XML Schema .xsd files and technical documentation are available on the Schema pages.
Migrating or sharing data with OME-XML¶
Data saved in an OME-XML file is easily read by any software capable of reading and interpreting OME-XML. OME software tools can export and import OME-XML files, but any OME-XML-aware software can be used.
In the figure Data Workflow, microscope image data is imported into My OME Database (more accurately, an instance of OME managing image data at one location). After a set of analyses is performed, this data is exported in its entirety (i.e. including binary image data, image metadata, and the associated analytic results) and then passed to a collaborator’s OME database (“Your OME Database”). This strategy packages analytic data together with the image data to ensure none of the critical interpretative information is lost.
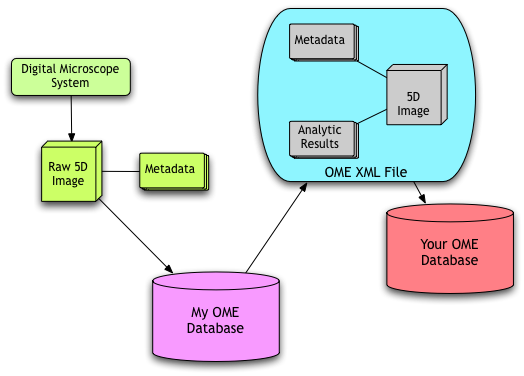
Data Workflow
Ongoing development¶
The ongoing development of the OME-XML data model can be tracked on https://trac.openmicroscopy.org/ome.
Users can also add work tickets to the system detailing any changes they feel should be made.