Note
This documentation is for OMERO 5.2. This version is now in maintenance mode and will only be updated in the event of critical bugs or security concerns. OMERO 5.3 is expected in the first quarter of 2017.
Local analysis¶
If you are interested in running your analysis locally and storing the results to the server, then your first step is to become familiar with the developer documentation.
- The Working with OMERO guide provides numerous examples in each language with explanations and tries to be a starting point for anyone who wants to write code which talks to the OMERO server.
- Most of the OMERO Application Programming Interface is covered by the Javadocs.
- Each of the languages has extra information on its own page:
Once you have your local analysis working, you can push it onto the server for background processing using the OMERO scripting service.
Storing external data in OMERO¶
There are several options for storing external or schema-less data in OMERO, including StructuredAnnotations for small quantities of data, or extending the OME model, but this risks interoperability issues. (See ExtendingOmero).
For larger volumes of data, or data which needs to be queried, OMERO.tables provides a unified solution for the storage of columnar data from various sources, such as automated analysis results or script-based processing, and makes them available within OMERO.
Third-party analysis and OMERO.tables¶
Support has been added for some third-party analysis data, which gets converted in OMERO into a common format. These formats include:
- MIAS data, measurements, and overlays
- InCell data and measurements
- Flex data with Acapella results (screencast). In the Flex case, additional configuration may be necessary for accessing both the raw data and the analysis results. Watch the configuration screencast for more information.
The analysis results which are parsed out of the formats listed above are converted to HDF by the OMERO.tables API. This facility can then be used by clients to visualize the parsed measurements, and in the case of regions of interest, see their location overlayed on the associated image:
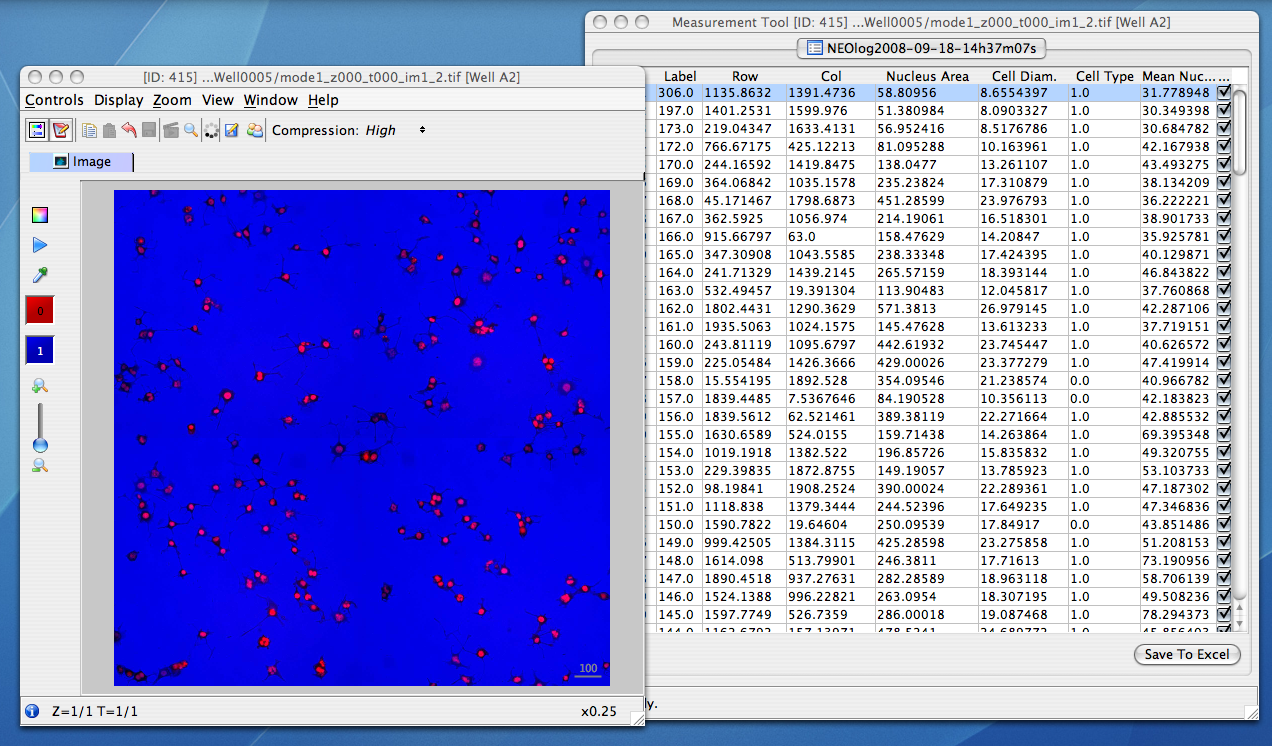
Other high-content screening (HCS) data¶
In addition to the Flex, Mias, and InCell 100 file formats, BD Pathway, Olympus ScanR, and native OME-XML/TIFF files can all be imported as HCS data, though without support for any external analysis data which may be attached. If you are interested in having other analysis formats supported, contact either the open source community or Glencoe Software, Inc. depending on your needs.