OME-TIFF specification¶
The following provides technical details on the OME-TIFF format. It assumes familiarity with both the TIFF specification and the OME Data Model, although there is some review of both.
The key words MUST, MUST NOT, REQUIRED, SHALL, SHALL NOT, SHOULD, SHOULD NOT, RECOMMENDED, MAY, and OPTIONAL in this specification are to be interpreted as described in RFC 2119.
An OME-TIFF dataset consists of:
- one or more files in standard TIFF format with the file extension
.ome.tifor.ome.tiffor BigTIFF format with one of these same file extensions or a BigTIFF-specific extension.ome.tf2,.ome.tf8or.ome.btf - a string of OME-XML metadata embedded in the ImageDescription tag of the first IFD (Image File Directory) of each file. The XML string must be UTF-8 encoded.
Note that BigTIFF-specific file extensions are an addition to the original specification, and software using an older version of the specification may not be able to handle these file extensions.
OME-XML metadata¶
This string is a standard block of OME-XML, except that instead of storing pixels as BinData elements with base64-encoded pixel data beneath them, it references pixels with TiffData elements that specify which TIFF IFD corresponds to each image plane. As such, the OME-XML string can be regarded as a “metadata block” because the actual pixels are stored within the TIFF structure, not the XML.
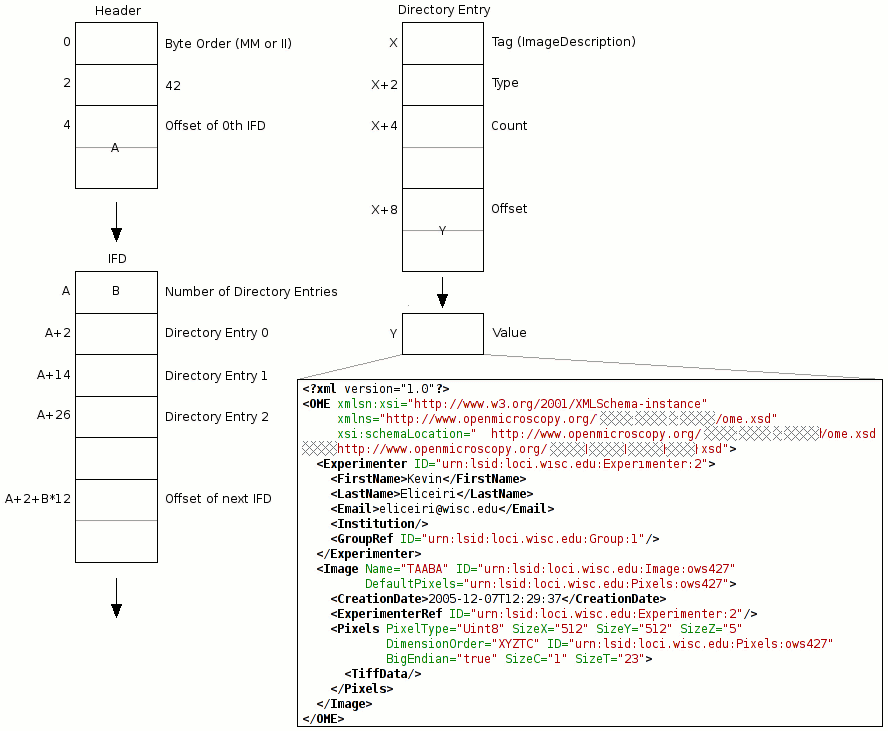
The diagram OME-TIFF header (adapted from the TIFF specification) shows the organization of a TIFF header along with the placement of the OME-XML metadata block. Note this is for the TIFF standard specification only; the header structure is slightly different for BigTIFF; see the BigTIFF file format specification. A TIFF file can contain any number of IFDs, with each one specifying an image plane along with certain accompanying metadata such as pixel dimensions, physical dimensions, bit depth, color table, etc. One of the fields an IFD can contain is ImageDescription, which provides a place to write a comment describing the corresponding image plane. This field is a convenient place to store the OME-XML metadata block—any TIFF library capable of parsing IFDs and extracting an ImageDescription comment can easily obtain an OME-TIFF file’s entire set of metadata as OME-XML.
Note
A TIFF file contains one IFD per image plane, but the OME-XML metadata block is stored only in the first IFD structure. However, for an image sequence distributed across multiple OME-TIFF files, each file will contain a copy of the OME-XML metadata block (see Partial OME-XML metadata below for exceptions to this rule). Thus, if any files are lost, the metadata is preserved. The OME-XML block in each file is nearly identical—the only difference is in the TiffData elements appearing beneath Pixels elements, since each TIFF file contains a different portion of the image data (see Multi-file OME-TIFF below).
DimensionOrder¶
As mentioned above, the standard OME-XML format encodes image planes as base64 character blocks contained within BinData elements beneath a Pixels element. The Pixels element has a DimensionOrder attribute that specifies the rasterization order of the image planes. For example, XYZTC means that there is a series of image planes with the Z axis varying fastest, followed by T, followed by C (e.g. if a XYZTC dataset contains two focal planes, three time points and two channels, the order would be: Z0-T0-C0, Z1-T0-C0, Z0-T1-C0, Z1-T1-C0, Z0-T2-C0, Z1-T2-C0, Z0-T0-C1, Z1-T0-C1, Z0-T1-C1, Z1-T1-C1, Z0-T2-C1, Z1-T2-C1).
Since a multi-page TIFF has no limit to the number of image planes it can contain, the same scheme described above for specifying the rasterization order works within the OME-TIFF file. The only difference is that instead of the pixels being encoded in base64 inside BinData elements, they are stored within the TIFF file in the standard fashion, one per IFD; see the TiffData element below for specifics.
TIFF comments¶
When embedding your OME-XML string as a TIFF comment, it is best practice to preface it with the following informative comment:
<!-- Warning: this comment is an OME-XML metadata block, which contains
crucial dimensional parameters and other important metadata. Please edit
cautiously (if at all), and back up the original data before doing so.
For more information, see the OME-TIFF documentation:
https://docs.openmicroscopy.org/latest/ome-model/ome-tiff/ -->
The TiffData element¶
As the illustration OME-TIFF header shows, all that is needed to indicate that the pixels are located within the enclosing TIFF structure is a TiffData element with no attributes. By default, the first IFD corresponds to the first image plane (Z0-T0-C0), and the rasterization order of subsequent IFDs is given by the Pixels element’s DimensionOrder attribute, as described above.
However, there are several attributes for TiffData elements allowing greater control over the dimensional position of each IFD:
- IFD - gives the IFD(s) for which this element is applicable. Indexed from 0. Default is 0 (the first IFD).
- FirstZ - gives the Z position of the image plane at the specified IFD. Indexed from 0. Default is 0 (the first Z position).
- FirstT - gives the T position of the image plane at the specified IFD. Indexed from 0. Default is 0 (the first T position).
- FirstC - gives the C position of the image plane at the specified IFD. Indexed from 0. Default is 0 (the first C position).
- PlaneCount - gives the number of IFDs affected. Dimension order of IFDs is given by the enclosing Pixels element’s DimensionOrder attribute. Default is the number of IFDs in the TIFF file, unless an IFD is specified, in which case the default is 1.
Here are some example XML fragments:
Fragment 1¶
<Pixels ID="urn:lsid:loci.wisc.edu:Pixels:ows325"
Type="uint8" DimensionOrder="XYZTC"
SizeX="512" SizeY="512" SizeZ="3" SizeT="2" SizeC="2">
<TiffData/>
</Pixels>
| IFD | Position |
|---|---|
| 0 | Z0-T0-C0 |
| 1 | Z1-T0-C0 |
| 2 | Z2-T0-C0 |
| 3 | Z0-T1-C0 |
| 4 | Z1-T1-C0 |
| 5 | Z2-T1-C0 |
| 6 | Z0-T0-C1 |
| 7 | Z1-T0-C1 |
| 8 | Z2-T0-C1 |
| 9 | Z0-T1-C1 |
| 10 | Z1-T1-C1 |
| 11 | Z2-T1-C1 |
The default TiffData tag simply assigns every IFD to a position according to the given DimensionOrder rasterization. If the TIFF file has more than SizeZ*SizeT*SizeC (3*2*2=12 in this case) IFDs, the remaining IFDs are extraneous and should be ignored by OME-TIFF readers.
Fragment 2¶
<Pixels ID="urn:lsid:loci.wisc.edu:Pixels:ows462"
Type="uint8" DimensionOrder="XYCTZ"
SizeX="512" SizeY="512" SizeZ="4" SizeT="3" SizeC="2">
<TiffData PlaneCount="10"/>
</Pixels>
| IFD | Position |
|---|---|
| 0 | Z0-T0-C0 |
| 1 | Z0-T0-C1 |
| 2 | Z0-T1-C0 |
| 3 | Z0-T1-C1 |
| 4 | Z0-T2-C0 |
| 5 | Z0-T2-C1 |
| 6 | Z1-T0-C0 |
| 7 | Z1-T0-C1 |
| 8 | Z1-T1-C0 |
| 9 | Z1-T1-C1 |
When specified, the PlaneCount attribute gives the number of IFDs affected by the TiffData element. In this case, even though the Pixels element defines 4*3*2=24 image planes total, the TiffData element assigns only 10 planes. The remaining 14 planes are unspecified and hence lost.
Fragment 3¶
<Pixels ID="urn:lsid:loci.wisc.edu:Pixels:ows197"
Type="uint8" DimensionOrder="XYZTC"
SizeX="512" SizeY="512" SizeZ="4" SizeC="3" SizeT="2">
<TiffData IFD="3" PlaneCount="5"/>
</Pixels>
| IFD | Position |
|---|---|
| 3 | Z0-T0-C0 |
| 4 | Z1-T0-C0 |
| 5 | Z2-T0-C0 |
| 6 | Z3-T0-C0 |
| 7 | Z0-T1-C0 |
States that the rasterization begins at the fourth IFD (IFD #3), and continues for five planes total. IFDs #0, #1 and #2 are not used, and should be ignored by OME-TIFF readers.
Fragment 4¶
<Pixels ID="urn:lsid:loci.wisc.edu:Pixels:ows789"
Type="uint8" DimensionOrder="XYZTC"
SizeX="512" SizeY="512" SizeZ="1" SizeC="1" SizeT="6">
<TiffData IFD="0" FirstT="5"/>
<TiffData IFD="1" FirstT="4"/>
<TiffData IFD="2" FirstT="3"/>
<TiffData IFD="3" FirstT="2"/>
<TiffData IFD="4" FirstT="1"/>
<TiffData IFD="5" FirstT="0"/>
</Pixels>
| IFD | Position |
|---|---|
| 0 | Z0-T5-C0 |
| 1 | Z0-T4-C0 |
| 2 | Z0-T3-C0 |
| 3 | Z0-T2-C0 |
| 4 | Z0-T1-C0 |
| 5 | Z0-T0-C0 |
Any number of TiffData elements may be provided. In this case, the dimensional positions of each of the first six IFDs are explicitly defined, effectively overriding the rasterization given by DimensionOrder, storing the planes in reverse temporal order.
For details on validating your OME-XML metadata block, see the validating OME-XML section on the Extracting, processing and validating OME-XML page.
Multi-file OME-TIFF¶
As demonstrated above, the OME-TIFF format is capable of storing the entire multidimensional image series in one master OME-TIFF file.
Alternatively, a collection of multiple OME-TIFF files may be used. Using the TiffData attributes outlined above together with UUID elements and attributes, the OME-XML metadata block can unambiguously define which dimensional positions correspond to which IFDs from which files. Each OME-TIFF need not contain the same number of images.
The only difference between the OME-XML metadata block per TIFF file is the UUID attribute of the root OME element. This value should be a distinct UUID value for each file, so that each TiffData element can unambiguously reference its relevant file using a UUID child element.
Note
The FileName attribute of the UUID is optional, but strongly recommended—otherwise, the OME-TIFF reader must scan OME-TIFF files in the working directory looking for matching UUID signatures.
When splitting an OME-TIFF across multiple files, the OME-XML metadata must either be embedded into each TIFF file or use partial metadata blocks.
Embedded OME-XML metadata¶
In the first case, a nearly identical OME-XML metadata block must be inserted into the first IFD of each constituent OME-TIFF file.
The only difference between the OME-XML metadata block per TIFF file is the UUID attribute of the root OME element. This value should be a distinct UUID value for each file, so that each TiffData element can unambiguously reference its relevant file using a UUID child element.
The 5D datasets demonstrate how OME-TIFF datasets can be distributed across multiple files. Each of the files in the set has identical metadata apart from the UUID, the unique identifier in each file. For example the identifiers could be distributed as follows:
tubhiswt_C0_TP0.ome.tif with ID 45c8bf18-6aa2-478c-9080-e0b0d3eb1f70
<OME xmlns="http://www.openmicroscopy.org/Schemas/OME/2016-06"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
Creator="OME Bio-Formats 5.2.0-m4"
UUID="urn:uuid:45c8bf18-6aa2-478c-9080-e0b0d3eb1f70"
xsi:schemaLocation="http://www.openmicroscopy.org/Schemas/OME/2016-06
http://www.openmicroscopy.org/Schemas/OME/2016-06/ome.xsd">
...
<Pixels BigEndian="false" DimensionOrder="XYZTC" ID="Pixels:0"
Interleaved="false" SignificantBits="8" SizeC="2" SizeT="43"
SizeX="512" SizeY="512" SizeZ="10" Type="uint8">
...
<TiffData FirstC="0" FirstT="0" FirstZ="0" IFD="0" PlaneCount="1">
<UUID FileName="tubhiswt_C0_TP0.ome.tif">
urn:uuid:45c8bf18-6aa2-478c-9080-e0b0d3eb1f70
</UUID>
</TiffData>
...
<TiffData FirstC="0" FirstT="1" FirstZ="1" IFD="1" PlaneCount="1">
<UUID FileName="tubhiswt_C0_TP1.ome.tif">
urn:uuid:743275b7-6726-46bd-b7bb-aca3085f429a
</UUID>
</TiffData>
...
tubhiswt_C0_TP1.ome.tif with ID 743275b7-6726-46bd-b7bb-aca3085f429a
<OME xmlns="http://www.openmicroscopy.org/Schemas/OME/2016-06"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
Creator="OME Bio-Formats 5.2.0-m4"
UUID="urn:uuid:743275b7-6726-46bd-b7bb-aca3085f429a"
xsi:schemaLocation="http://www.openmicroscopy.org/Schemas/OME/2016-06
http://www.openmicroscopy.org/Schemas/OME/2016-06/ome.xsd"
...
<Pixels BigEndian="false" DimensionOrder="XYZTC" ID="Pixels:0"
Interleaved="false" SignificantBits="8" SizeC="2" SizeT="43"
SizeX="512" SizeY="512" SizeZ="10" Type="uint8">
...
<TiffData FirstC="0" FirstT="0" FirstZ="0" IFD="0" PlaneCount="1">
<UUID FileName="tubhiswt_C0_TP0.ome.tif">
urn:uuid:45c8bf18-6aa2-478c-9080-e0b0d3eb1f70
</UUID>
</TiffData>
...
<TiffData FirstC="0" FirstT="1" FirstZ="0" IFD="0" PlaneCount="1">
<UUID FileName="tubhiswt_C0_TP1.ome.tif">
urn:uuid:743275b7-6726-46bd-b7bb-aca3085f429a
</UUID>
</TiffData>
...
And so on for files:
- tubhiswt_C0_TP2.ome.tif with ID 1f462b60-b508-446e-b42a-c4e6fa2a44e8
- tubhiswt_C0_TP3.ome.tif with ID a023901d-43fd-44f2-b4be-159afa1e985c
- …
Partial OME-XML metadata¶
Instead of embedding the full OME-XML metadata into the header of each OME-TIFF, partial OME-XML metadata blocks can be stored in some or all of the OME-TIFF files using the Binary-Only element as illustrated below:
<?xml version="1.0" encoding="UTF-8"?>
<OME UUID="urn:uuid:4978087c-a670-4b12-af53-256c62d8d101"
xmlns="http://www.openmicroscopy.org/Schemas/OME/2016-06"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:schemaLocation="http://www.openmicroscopy.org/Schemas/OME/2016-06
http://www.openmicroscopy.org/Schemas/OME/2016-06/ome.xsd">
<BinaryOnly MetadataFile="multifile.companion.ome"
UUID="urn:uuid:07504f88-7bc3-11e0-b937-2faf67bc00b3"/>
</OME>
The MetadataFile element should contain the name of the master file containing the full OME-XML metadata block and UUID should contain the UUID of this master file.
The master file containing the full OME-XML metadata should be either an
OME-XML companion file with the extension .companion.ome or a master
OME-TIFF file containing the full metadata (see Multi-file OME-TIFF filesets for
representative samples).
Sub-resolutions¶
New in version 6.0.0.
OME-TIFF supports multi-resolution images or pyramidal images where individual planes are stored at different levels of resolution. The downsampled image planes are called pyramidal levels, sub-resolution image planes or sub-resolutions.
Supported resolutions¶
OME-TIFF planes can be reduced along the X and Y dimensions. Each pyramidal level must be a downsampling of the full-resolution plane in the X and Y dimensions and the resolution should stay unchanged in the other dimensions. The downsampling factor:
- should be an integer value,
- should be identical along the X and the Y dimensions,
- should stay the same between each consecutive pyramid level.
The following table below shows two examples of pyramid level dimensions using typical downsampling factors:
| Example 1 (X × Y × Z × C × T) | Example 2 (X × Y × Z × C × T) | |
|---|---|---|
| Downsampling factor | 3 | 4 |
| Level 0 (full-resolution) | 9234 × 6075 × 1 × 1 × 10 | 38912 × 25600 × 200 × 3 × 1 |
| Level 1 | 3078 × 2025 × 1 × 1 × 10 | 9728 × 6400 × 200 × 3 × 1 |
| Level 2 | 1026 × 675 × 1 × 1 × 10 | 2432 × 1600 × 200 × 3 × 1 |
| Level 3 | 342 × 225 × 1 × 1 × 10 | 608 × 400 × 200 × 3 × 1 |
| Level 4 | 114 × 74 × 1 × 1 × 10 | 152 × 100 × 200 × 3 × 1 |
| Level 5 | 38 × 25 × 1 × 1 × 10 |
Storage¶
Full-resolution image planes must remain stored as described above using a valid TIFF IFD and referenced from the OME-XML metadata using the TiffData element.
Each sub-resolution must be stored in the same IFD as the full-resolution image plane. Additionally:
- the offsets of all sub-resolutions IFDs must be referenced from the IFD of the full-resolution plane using the SubIFDs TIFF extension tag 330 as defined in the TIFF Tech Note 1 of the Adobe PageMaker® 6.0 TIFF Technical Notes. The list of sub-resolution offsets must be ordered by plane size from largest to smallest,
- the IFD offsets of pyramidal levels must neither be referenced in the primary chain of IFDs derived from the first IFD of the TIFF file nor be referenced in a TiffData element of the OME-XML metadata,
- the NewSubFileType TIFF tag 254 for each pyramidal level should be set to 1 to distinguish full-resolution planes from downsampled planes.
The planes of largest resolutions should be organized into tiles rather than strips as described in the TIFF specification and may be compressed using any of the officially supported schemes including LZW, JPEG or JPEG2000. Sub-resolution image planes may choose to use different compression algorithms than the one used by the full resolution plane. For example the full resolution image may use no compression or lossless compression while the sub-resolution images use lossy compression.
While baseline TIFF may suffice for smaller pyramidal images, BigTIFF is recommended for large images.
See also
- https://openmicroscopy.github.io/design/OME005/
- Official design proposal for the addition of sub-resolution support to the OME-TIFF specification.