Compliant file specification¶
This was developed in conjunction with the April 2010 release of the OME-XML Model and the samples have been updated to the June 2016 release.
A “compliant” specification OME file has been defined. This is not the minimum required for the display of an image (for this, see Minimum Specification). This is the information a file should contain to authoritatively describe an imaging experiment, so that another person could, with the same sample and microscope, reproduce the data recorded in the file. Therefore, an ‘OME Compliant’ file should be as complete as possible, but only contain metadata relevant to a specific imaging experiment i.e. all of the fields listed below that are relevant to the experiment should be completed. As an example, PockelCellSetting would not be used in most wide-field microscopy experiments, but would be mandatory for many multi-photon imaging experiments.
Sample structure¶
Two samples of this structure are shown below. They were generated by opening a propriety file (Olympus .oib and DeltaVision .dv, respectively) in ImageJ, using the Bio-Formats plugin with the Display OME-XML Metadata option checked. A few additional attributes were then manually added to the files to make them compliant.
Sample structure one:
<?xml version="1.0"?>
<OME xmlns="http://www.openmicroscopy.org/Schemas/OME/2016-06"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:str="http://exslt.org/strings"
xsi:schemaLocation="http://www.openmicroscopy.org/Schemas/OME/2016-06 http://www.openmicroscopy.org/Schemas/OME/2016-06/ome.xsd">
<Image ID="Image:0" Name="Series 1">
<AcquisitionDate>2008-02-06T13:43:19</AcquisitionDate>
<Description>An example OME compliant file, based on Olympus.oib</Description>
<Pixels DimensionOrder="XYCZT" ID="Pixels:0" PhysicalSizeX="0.207" PhysicalSizeY="0.207" SizeC="3" SizeT="16" SizeX="1024" SizeY="1024" SizeZ="1" TimeIncrement="120.1302" Type="uint16">
<Channel EmissionWavelength="523" ExcitationWavelength="488" ID="Channel:0:0" IlluminationType="Epifluorescence" Name="CH1" SamplesPerPixel="1" PinholeSize="103.5" AcquisitionMode="LaserScanningConfocalMicroscopy"/>
<Channel EmissionWavelength="578" ExcitationWavelength="561" ID="Channel:0:1" IlluminationType="Epifluorescence" Name="CH3" SamplesPerPixel="1" PinholeSize="127.24" AcquisitionMode="LaserScanningConfocalMicroscopy"/>
<Channel ExcitationWavelength="488" ID="Channel:0:2" IlluminationType="Transmitted" ContrastMethod="DIC" Name="TD1" SamplesPerPixel="1" AcquisitionMode="LaserScanningConfocalMicroscopy"/>
<BinData BigEndian="false" Length="0"/>
</Pixels>
</Image>
</OME>
Sample structure two:
<?xml version="1.0"?>
<OME xmlns="http://www.openmicroscopy.org/Schemas/OME/2016-06"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:str="http://exslt.org/strings"
xsi:schemaLocation="http://www.openmicroscopy.org/Schemas/OME/2016-06 http://www.openmicroscopy.org/Schemas/OME/2016-06/ome.xsd">
<Instrument ID="Instrument:0">
<Detector ID="Detector:0:0" Model="COOLSNAP_HQ / ICX285" Type="CCD"/>
<Objective ID="Objective:10002" Immersion="Oil" LensNA="1.4" Manufacturer="Olympus" NominalMagnification="100"/>
</Instrument>
<Image ID="Image:1" Name="example_R3D_D3D.dv">
<AcquisitionDate>2005-01-28T13:50:08</AcquisitionDate>
<Description>An example OME compliant file,
based on a wide-field microscope image</Description>
<ObjectiveSettings ID="Objective:10002" Medium="Oil" RefractiveIndex="1.52" CorrectionCollar="7"/>
<Pixels DimensionOrder="XYCZT" ID="Pixels:1" PhysicalSizeX="0.06631" PhysicalSizeY="0.06631" PhysicalSizeZ="0.2" SizeC="3" SizeT="1" SizeX="480" SizeY="480" SizeZ="5" Type="int16">
<Channel EmissionWavelength="457" ExcitationWavelength="360" ID="Channel:1:0" NDFilter="0.5" Name="DAPI" SamplesPerPixel="1" Fluor="DAPI" IlluminationType="Epifluorescence" ContrastMethod="Fluorescence" AcquisitionMode="WideField" Color="65535">
<DetectorSettings Binning="1x1" Gain="0.5" ID="Detector:0:0" ReadOutRate="10.0"/>
</Channel>
<Channel EmissionWavelength="528" ExcitationWavelength="490" ID="Channel:1:1" NDFilter="0.0" Name="FITC" SamplesPerPixel="1" Fluor="GFP" IlluminationType="Epifluorescence" ContrastMethod="Fluorescence" AcquisitionMode="WideField" Color="16711935">
<DetectorSettings Binning="1x1" Gain="0.5" ID="Detector:0:0" ReadOutRate="10.0"/>
</Channel>
<Channel EmissionWavelength="617" ExcitationWavelength="555" ID="Channel:1:2" NDFilter="0.0" Name="RD-TR-PE" SamplesPerPixel="1" Fluor="TRITC" IlluminationType="Epifluorescence" ContrastMethod="Fluorescence" AcquisitionMode="WideField" Color="-16776961">
<DetectorSettings Binning="1x1" Gain="0.5" ID="Detector:0:0" ReadOutRate="10.0"/>
</Channel>
<BinData BigEndian="false" Length="0"/>
<Plane DeltaT="0.0" ExposureTime="0.1" PositionX="3316.37" PositionY="-646.46" PositionZ="-21.496" TheC="0" TheT="0" TheZ="0"/>
<Plane DeltaT="0.294" ExposureTime="0.1" PositionX="3316.37" PositionY="-646.46" PositionZ="-21.696" TheC="0" TheT="0" TheZ="1"/>
<Plane DeltaT="0.587" ExposureTime="0.1" PositionX="3316.37" PositionY="-646.46" PositionZ="-21.896" TheC="0" TheT="0" TheZ="2"/>
<Plane DeltaT="0.881" ExposureTime="0.1" PositionX="3316.37" PositionY="-646.46" PositionZ="-22.096" TheC="0" TheT="0" TheZ="3"/>
<Plane DeltaT="1.174" ExposureTime="0.1" PositionX="3316.37" PositionY="-646.46" PositionZ="-22.296" TheC="0" TheT="0" TheZ="4"/>
<Plane DeltaT="9.625" ExposureTime="0.3" PositionX="3316.37" PositionY="-646.46" PositionZ="-21.496" TheC="1" TheT="0" TheZ="0"/>
<Plane DeltaT="10.12" ExposureTime="0.3" PositionX="3316.37" PositionY="-646.46" PositionZ="-21.696" TheC="1" TheT="0" TheZ="1"/>
<Plane DeltaT="10.613" ExposureTime="0.3" PositionX="3316.37" PositionY="-646.46" PositionZ="-21.896" TheC="1" TheT="0" TheZ="2"/>
<Plane DeltaT="11.106" ExposureTime="0.3" PositionX="3316.37" PositionY="-646.46" PositionZ="-22.096" TheC="1" TheT="0" TheZ="3"/>
<Plane DeltaT="11.599" ExposureTime="0.3" PositionX="3316.37" PositionY="-646.46" PositionZ="-22.296" TheC="1" TheT="0" TheZ="4"/>
<Plane DeltaT="25.447" ExposureTime="0.1" PositionX="3316.37" PositionY="-646.46" PositionZ="-21.496" TheC="2" TheT="0" TheZ="0"/>
<Plane DeltaT="25.739" ExposureTime="0.1" PositionX="3316.37" PositionY="-646.46" PositionZ="-21.696" TheC="2" TheT="0" TheZ="1"/>
<Plane DeltaT="26.033" ExposureTime="0.1" PositionX="3316.37" PositionY="-646.46" PositionZ="-21.896" TheC="2" TheT="0" TheZ="2"/>
<Plane DeltaT="26.326" ExposureTime="0.1" PositionX="3316.37" PositionY="-646.46" PositionZ="-22.096" TheC="2" TheT="0" TheZ="3"/>
<Plane DeltaT="26.619" ExposureTime="0.1" PositionX="3316.37" PositionY="-646.46" PositionZ="-22.296" TheC="2" TheT="0" TheZ="4"/>
</Pixels>
</Image>
</OME>
Note
The data (BinData element content) in these sample was removed for reasons of length.
Alternative valid forms:
- would have a TiffData block instead of the BinData block (this would be used in the header of an OME-TIFF file)
- would have a MetadataOnly block instead of the BinData block (this would be used as a companion to one or more BinaryOnly OME-TIFF files)
Note
A units system was added in January 2015. This sample assumes the default unit for each value is used.
Definitions of values stored¶
The figure Definitions of values stored is ©2010 Linkert et al. Figure originally published as Figure 2, Linkert et al (2010), J. Cell Biol. 189(5):777-782 (http://jcb.rupress.org/content/189/5/777)
Note
AcquiredDate was renamed to AcquisitionDate in June 2012
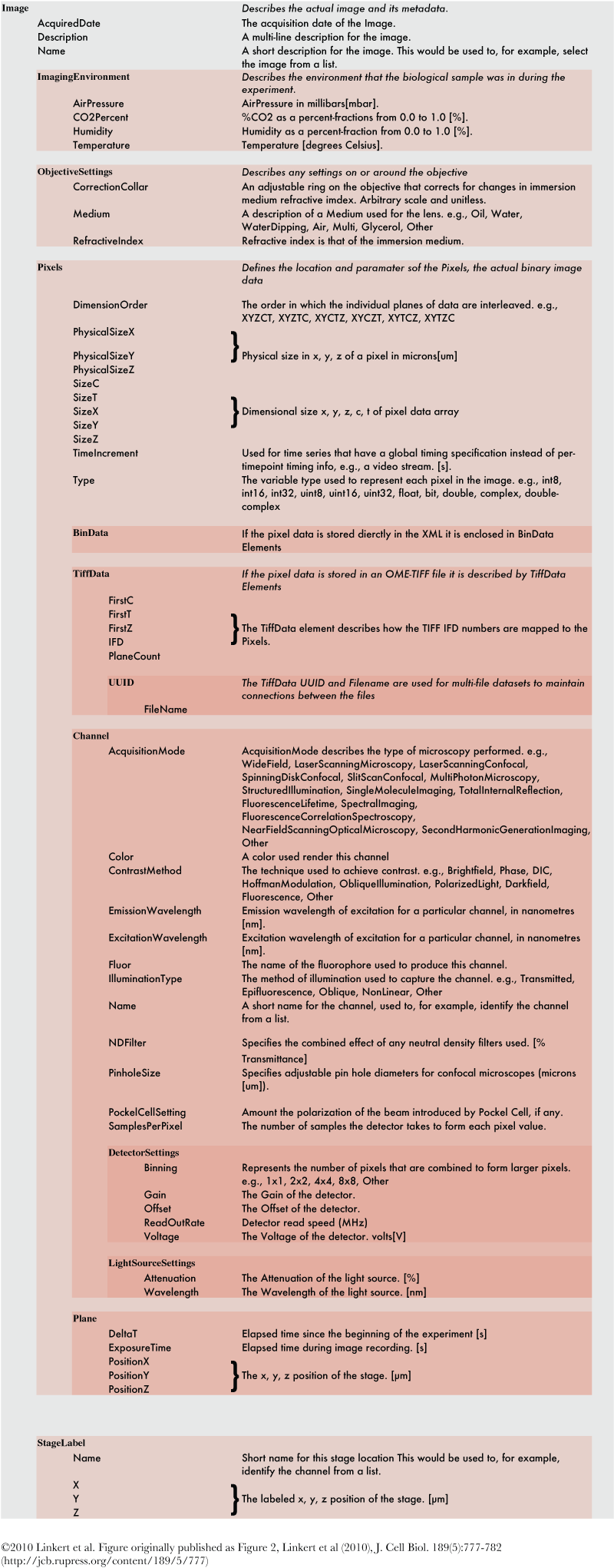
Definitions of values stored